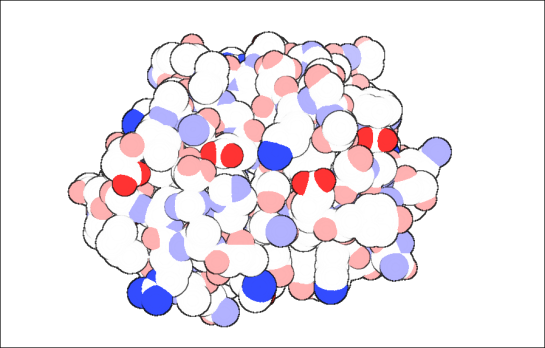
Almost all of us visiting PDB would have looked at the image shown below that attract us like moths attracted to a light. I am talking about the aesthetically pleasing protein images created by David Goodsell.

p53 Tumor Suppresor.
Image Courtesy: http://dx.doi.org/10.2210/rcsb_pdb/mom_2002_7
In case you didn’t know, he is the author of Molecule of the Month series. Since the images look anything like the ones we usually keep looking at, one is attracted to the level of abstraction the image projects due to which, the reader understands the big picture. And, of course, they come in all cute colors!
To quote him about the artwork’s intention, that is to give
a pictorial overview of the molecules that orchestrate the process of life. [2]
Some history about David Goodsell can be found in his website and here.
So, you have correctly guessed that this post is about how to make such “David Goodsell-sque” images of your protein. For consistency, I used 2BEM, a CBM33 polysaccharide monooxygenase enzyme [3]. The view is looking at the protein’s active site, which is quite planar.
Using Python Molecule Viewer
PMV [4] is good tool to run AutoDock, and manipulating structures. This link explains in basically two steps of getting such images. Here is my try with PMV
Using PyMOL
Can PyMOL be far behind in implementing this? I didn’t think so. Fortunately, there are at least two alternate ways of doing this in PyMOL.
1. Using a couple of commands. After loading the protein, type in the following commands as given here [5]
- unset specular
- set ray_trace_gain, 0
- set ray_trace_mode, 3
- bg_color white
- set ray_trace_color, black
- unset depth_cue
- ray
In surface,
If you want to change the colors to David Goodsell-sque, then it comes out like this. The important commands are the “specular” and “ray_trace_mode, 3”.
2. Are you thinking “Meh!, Close enough. But, not what I was looking for.”? Then, with some small tweaking in PyMOL you could get like this, using GLSL shaders [6, 7]
If you are using VMD version 1.8.7 or 1.8.6, then you can follow this link [8]
After loading the molecule, this is what I did to get the following image
- Main Menu->Graphics-> Representations
- Coloring method->Element
- Drawing Method->VDW
- Material->Goodsell
- Main Menu->Display->Rendermode->GLSL
- Main Menu->Display->Light 3
- Main Menu->Graphics-Materials->Goodsell->I played around with the diff parameters given under that
- Main Menu->Graphics->Colors->Categories->Element->C->white
- Main Menu->Display->Axes->Off
Using VMD and Blender
In case, you need to make such images to make a great impression, you can learn how to use Blender. It is a open-source 3D tool that can create models and has huge applications in animated movies, interior designing and other non-science stuff. See this link for more details: http://chemistry.stackexchange.com/questions/484/what-software-is-used-to-generate-the-pdb-molecule-of-the-month-images
I did not use VMD or Blender for 2BEM so using one of the images given in the above link. [9]
Using QuteMol
This is probably the ONE-CLICK David-Goodsell-sque image making tool. Download it from here. After loading the molecule, just click on the “Molecule of the Month” button. Voila!
A downside was I was unable to change the carbons to white. Sigh!
Anyway, I hope you had fun making images like I did. Enjoy! 🙂
References:
- http://mgl.scripps.edu/people/goodsell/
- http://www.asbmb.org/asbmbtoday/asbmbtoday_article.aspx?id=13702&page_id=2
- http://dx.doi.org/10.1074/jbc.M407175200
- Sanner MF (1999). Python: a programming language for software integration and development. Journal of molecular graphics & modelling, 17 (1), 57-61 PMID: 10660911
- http://www.pymolwiki.org/index.php/Gallery
- http://pymolwiki.org/index.php/GLSL_Shaders
- http://kpwu.wordpress.com/2012/06/28/pymol-draw-goodsell-like-view-using-glsl/
- http://www.ks.uiuc.edu/Research/vmd/minitutorials/glsloutline/
- http://chemistry.stackexchange.com/questions/484/what-software-is-used-to-generate-the-pdb-molecule-of-the-month-images
- http://pmvbase.blogspot.com/2010/04/making-david-goodsell-like.html
- http://www.researchgate.net/post/How_can_I_draw_proteins_like_in_PDB_Molecules_of_the_Month
- http://biostumblematic.wordpress.com/2009/12/02/rendering-proteins-in-pymol/
- Sanner MF, Olson AJ, & Spehner JC (1996). Reduced surface: an efficient way to compute molecular surfaces. Biopolymers, 38 (3), 305-20 PMID: 8906967
- Goodsell, D. (2002). p53 RCSB Protein Data Bank DOI: 10.2210/rcsb_pdb/mom_2002_7